Research Interests:
My research is part of the larger effort in the David D. Thomas lab on the study of structural dynamics associated with force-generating proteins during muscle contraction and the molecular mechanisms of catalysis and regulation of the Ca-ATPase (SERCA) that pumps calcium into the sarcoplasmic reticulum and thus relaxes the muscle. My goal is specifically, to use molecular modeling and molecular dynamics simulations to aid the interpretation of experimental results and to generate new structural and dynamical models that describe the function of muscle proteins on an atomistic level.
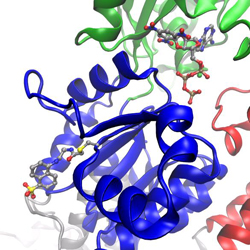
I work on many different projects, the main project currently is to use molecular dynamics simulations on SERCA labeled with the fluorescent probe IAEDANS with the aim to establish a more rigorous foundation for analyzing spectroscopic data. Site-specific labeling of a protein with a fluorescent probe can provide insight into local structural dynamics, based on fluorescence quenching or anisotropy measurements, or used to measure distances based on fluorescence resonance energy transfer (FRET) to another label. SERCA was labeled at position Cys674 in the P-domain (shown blue in figure) with the fluorescent probe IAEDANS. The starting point for the molecular dynamics simulations of the fluorescent probe and its protein environment was a new crystal structure of IAEDANS-labeled SERCA that was determined in collaboration with Howard Young, Univ. of Alberta, Edmonton. The structure was determined to 3.4 resolution, which was sufficient to show the IAEDANS label in close proximity to residues Arg615 and Arg620. To be able to perform these simulations, we developed CHARMM force-field parameters for the fluorescence probe IAEDANS. Quantum chemistry calculations have also been performed on the ground state and excited states of IAEDANS, to determine the orientation of the transition dipole moment. The transition dipole autocorrelation functions and reorientation times were calculated from the simulated trajectories and compared with experimental measurements by fluorescence anisotropy. FRET parameters were also determined from the simulations and compared with results from fluorescence experiments using IAEDANS as the donor and TNP-ADP bound in the nucleotide pocket as the acceptor. The results show that we have established a reliable framework for both fluorescence experiments and MD simulations in this system.
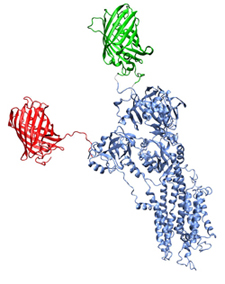
Another project is the simulation of XFP-SERCA fusion proteins to compare with experimental data from FRET. This project is a collaboration with Seth Robia, Loyolla University, Chicago. An ensemble of conformations of the XFP fused to SERCA is generated using a Monte-Carlo based method. From that ensemble of structures we can calculate FRET parameters using both distance distributions and orientation information and compare that to the experimental data. The aim is to understand the dynamics i.e. conformational changes in the cytoplasmic domains in SERCA.
Support:
My current work has been supported by NIH (GM27906, AR007612) and the Minnesota Supercomputing Institute.
Research Experience:
I have mostly been doing theoretical research on protein structure and function, but have significant laboratory experience as well in the field of biophysics/spectroscopy and biochemistry. Before the current work on muscle proteins I have worked on the opioid receptors and GPCR's, and on the photosynthetic proteins involved in oxygen evolution i.e. photosystem II.
The theoretical techniques I have experience with include molecular modeling of proteins, molecular dynamics simulations, quantum chemistry calculations of protein ligands and spectroscopic probes. I have also used bioinformatics tools to identify important residues and regions of protein sequences based on multiple sequence alignments.
The experimental measurement techniques I have used include EPR, FTIR, CD, fluorescence, UV/Vis, thermoluminescence, SAXS, mass-spectroscopy, steady state and flash oxygen polarography etc. I was member of a team involved in the development of kinetic fluorescence and oxygen polarography instruments at UIUC. The instrument we designed were commercially available through Artisan Scientific, Champaign, IL
Software I use include, CHARMM, CHARMM-GUI, MMTSB toolset, VMD, DS Visualizer, OriginPro, ProFit, CE (Combinatorial Extension), Gaussian/Gaussview, Molden, ConQuest, Situs, FPMOD, pyMol, DOWSER, Reduce, Procheck, ClustalW,EMBOSS.
Other Skills:
System administration experience on Linux (including setting up a small HPC cluster), UNIX (SGI), and Microsoft Windows environments. Computer hardware maintenance, repairs, and upgrades. Programming in Fortran, C, and scripting languages Perl, C-shell. User interface programming in Perl/Tk.
Education:
B.S. Chemistry & Microbiology, Stockholm University, Sweden, 1990
Ph.D. Biochemistry, Stockholm University, Sweden (S. Styring) 1996
Post Doc:
Center for Biophysics and Computational Biology and Dept. of Microbiology, University of Illinois (A. Crofts) 1996-99
Dept. of Biochemistry, Molecular Biology and Biophysics, University of Minnesota (B. Barry) 1999-02
Dept. of Medicinal Chemistry, University of Minnesota (D. Ferguson) 2002-07
Dept. of Biochemistry, Molecular Biology and Biophysics, University of Minnesota (D.D. Thomas) 2007-present
Recent Publications:
25. A bifunctional spin label reports the structural topology of phospholamban in magnetically-aligned bicelles. (2016) McCaffrey J.E., James Z.M., Svensson B., Binder B.P., Thomas D.D. J. Magn. Reson. 262, 50-6.
24. Molecular modeling of fluorescent SERCA biosensors. (2016) Svensson B., Autry J.M., Thomas D.D. Methods Mol. Biol. 1377, 503-22.
23. Bifunctional spin labeling of muscle proteins: accurate rotational dynamics, orientation, and distance by EPR. (2015) Thompson A.R., Binder B.P., McCaffrey J.E., Svensson B., Thomas D.D. Methods Enzymol. 2015;564:101-23.
22. FRET-based trilateration of probes bound within functional ryanodine receptors. (2014) Svensson B., Oda T., Nitu F.R., Yang Y., Cornea I., Chen-Izu Y., Fessenden J.D., Bers D.M., Thomas D.D., Cornea R.L. Biophys. J. 107, 2037-48. (*Cover article)
21. Structural mapping of divergent regions in the type 1 ryanodine receptor using fluorescence resonance energy transfer.(2014) Mahalingam M., Girgenrath T., Svensson B., Thomas D.D., Cornea R.L., Fessenden J.D. Structure 22, 322-32.
20. Nucleotide activation of the Ca-ATPase. (2012) Autry J.M., Rubin J.E., Svensson B., Li J., Thomas D.D. J. Biol. Chem. 287, 39070-82.
19. Lin, A. Y., E. Prochniewicz, Z. M. James, B. Svensson, and D. D. Thomas (2011) "Large-scale opening of utrophin's tandem CH domains upon actin binding, by an induced-fit mechanism" Proc. Natl. Acad. Sci. U.S.A., 108: 12729-12733.
18. Rose, S., Minagawa, J., Seufferheld, M., Padden, S., Svensson, B., Kolling, D.R.J., Crofts, A.R. and Govindjee (2008) "D1-arginine257 mutants (R257E, K, and Q) of Chlamydomonas reinhardtii have a lowered QB redox potential: analysis of thermoluminescence and fluorescence measurements", Photosynth. Res. 98, 449-468.
17. Klein, J.C., Burr, A.R., Svensson, B., Kennedy, D.J., Allingham, J., Titus, M.A., Rayment, I. and Thomas, D.D. (2008) "Actin-binding cleft closure in myosin II probed by site-directed spin labeling and pulsed EPR", Proc. Natl. Acad. Sci. U.S.A. 105, 12867-12872.
16. Winters, D.L., Autry, J.M., Svensson, B., and Thomas, D.D. (2008) "Interdomain fluorescence resonance energy transfer in SERCA probed by cyan-fluorescent protein fused to the actuator domain", Biochemistry 47, 4246-4256.
15. Goodell, J.R., Svensson, B., and Ferguson, D.M. (2006) "Spectrophotometric Determination and Computational Evaluation of the Rates of hydrolysis of 9-Amino-Substituted Acridines", J. Chem. Inf. Model. 46, 876-883.